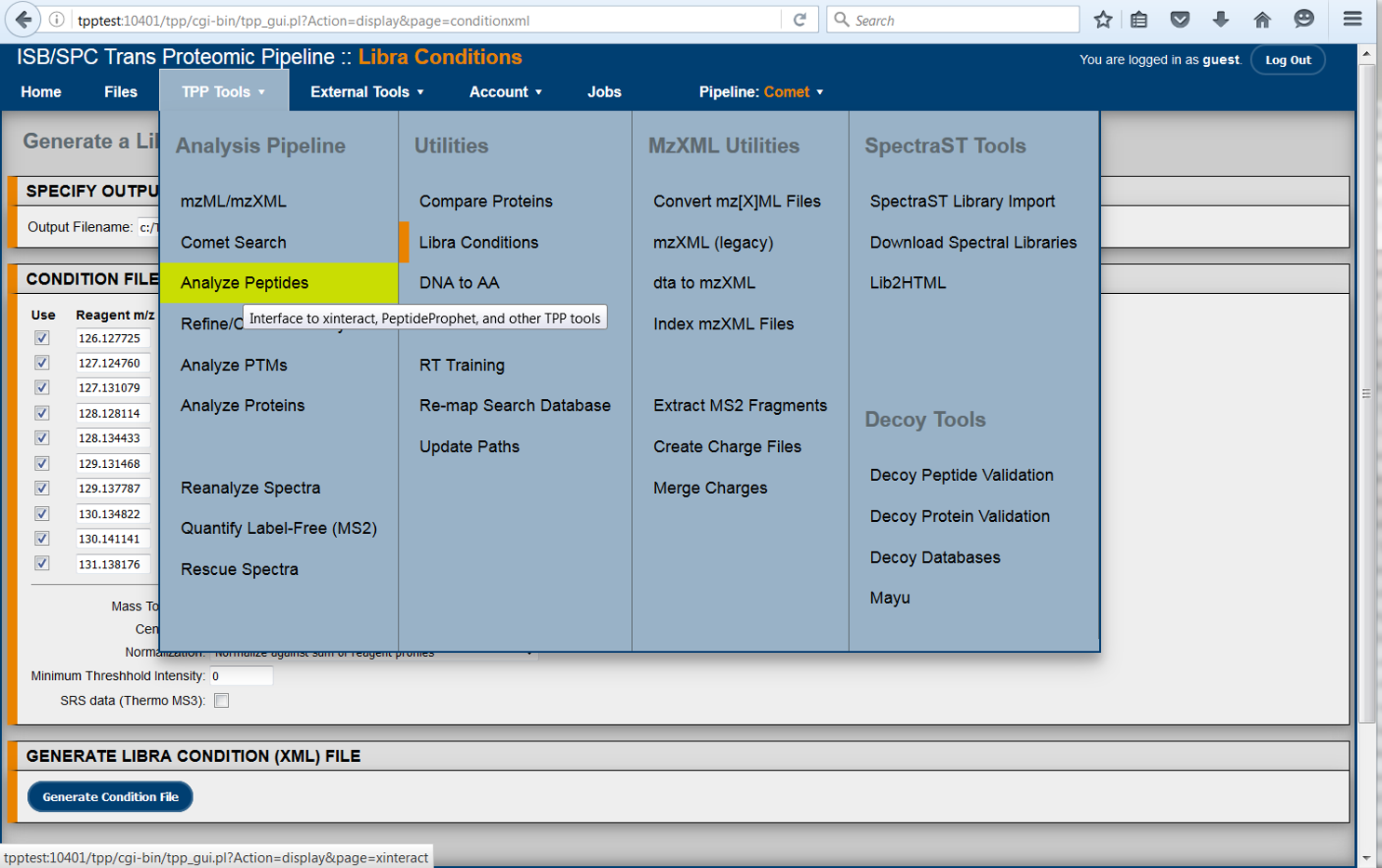
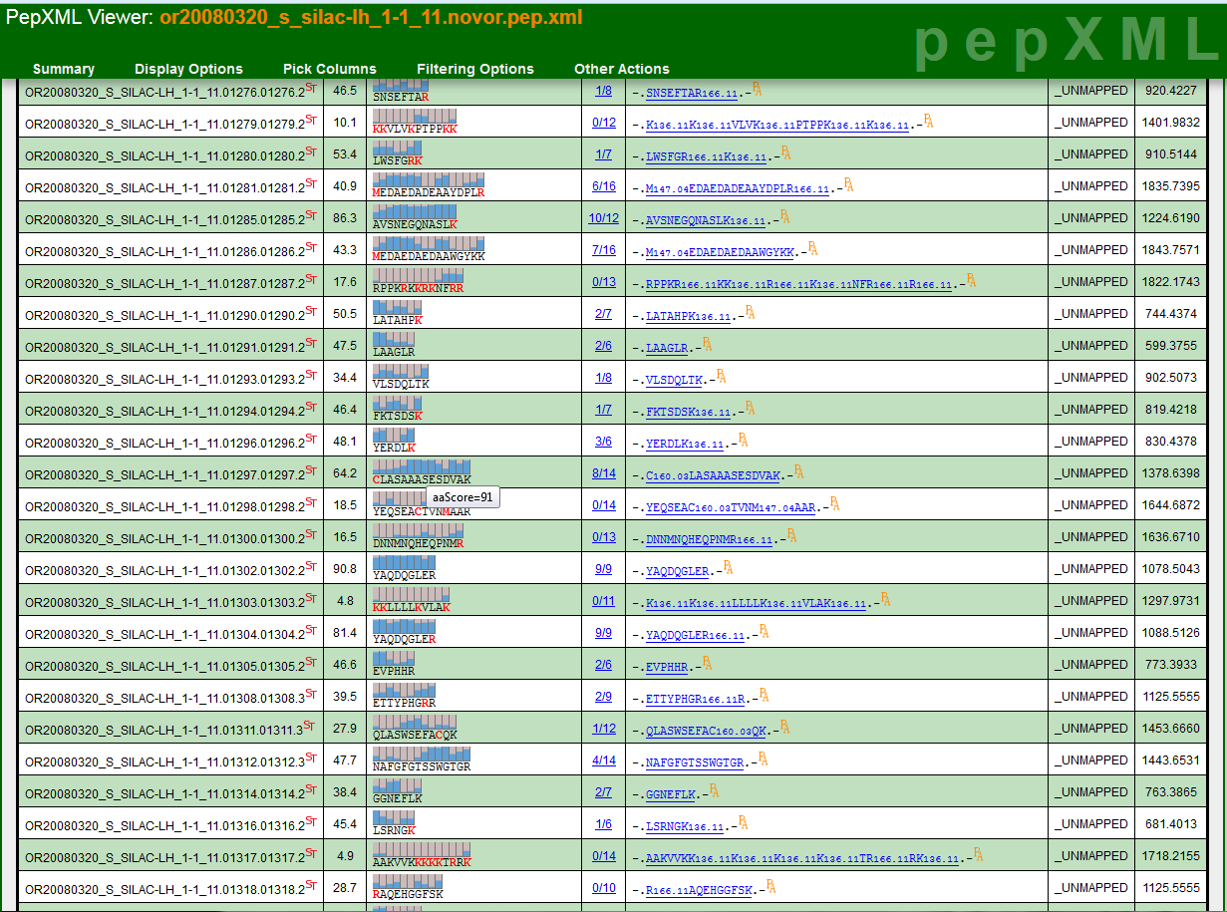
The TPP is a complete and mature suite of free and open-source software tools for MS data representation, MS data visualization, peptide identification and validation, protein identification, quantification, and annotation, data storage and mining, and biological inference.
The TPP is available on Linux and Windows systems and builds on Mac OS X as well, and supports a well-established worldwide user community.
The latest TPP version is 7.3.0 (Trade Wind), released April 2025.
Read the Release Notes and Installation Notes for the latest version.
Download directly from Sourceforge.
Trans-Proteomic Pipeline: Robust Mass Spectrometry-Based Proteomics Data Analysis Suite
Eric W. Deutsch, Luis Mendoza, David D. Shteynberg, Michael R. Hoopmann, Zhi Sun, Jimmy K. Eng, and Robert L. Moritz
Journal of Proteome Research 2023 22 (2), 615-624
DOI: 10.1021/acs.jproteome.2c00624
To cite other TPP components, please refer to this page.
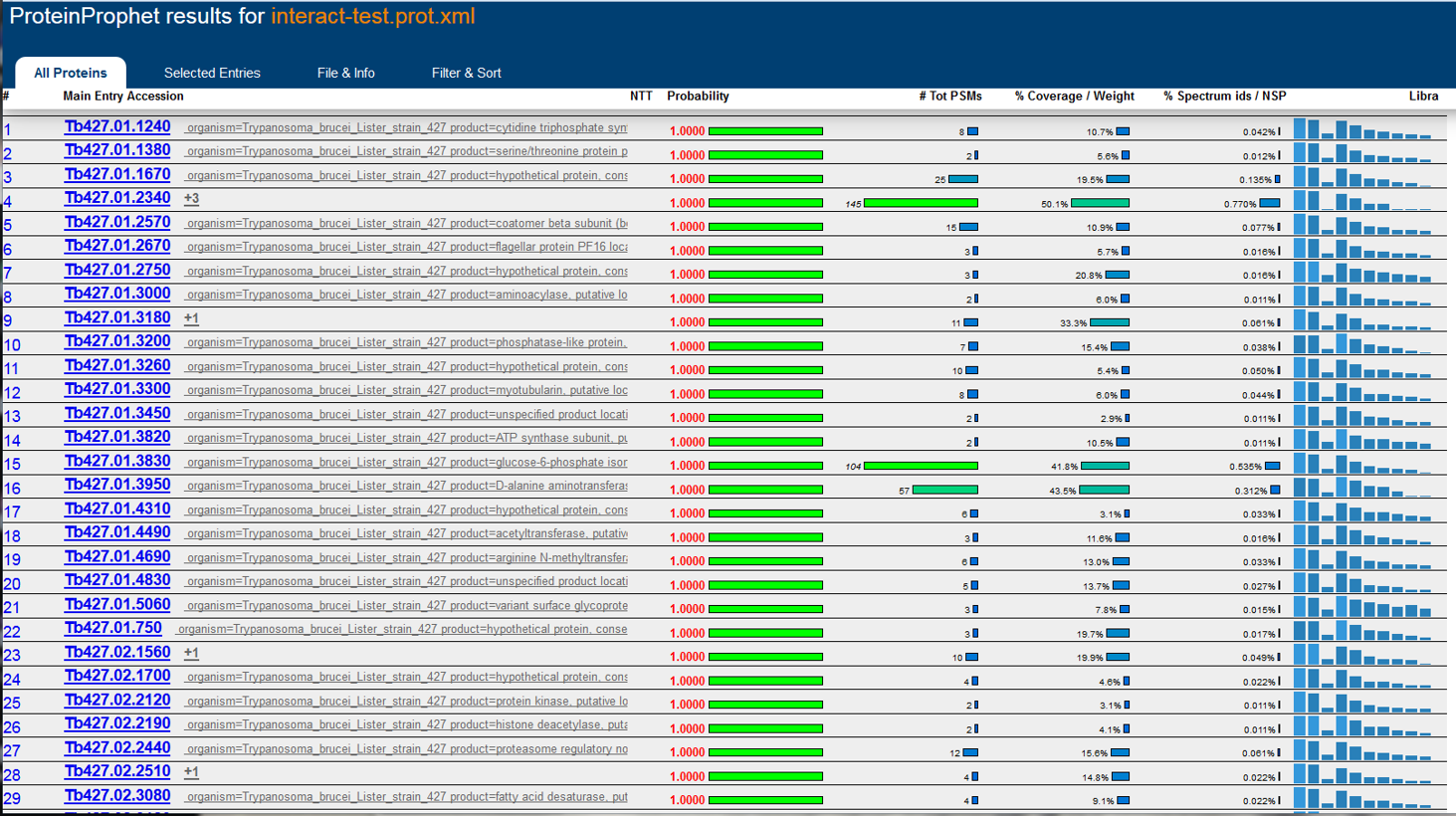
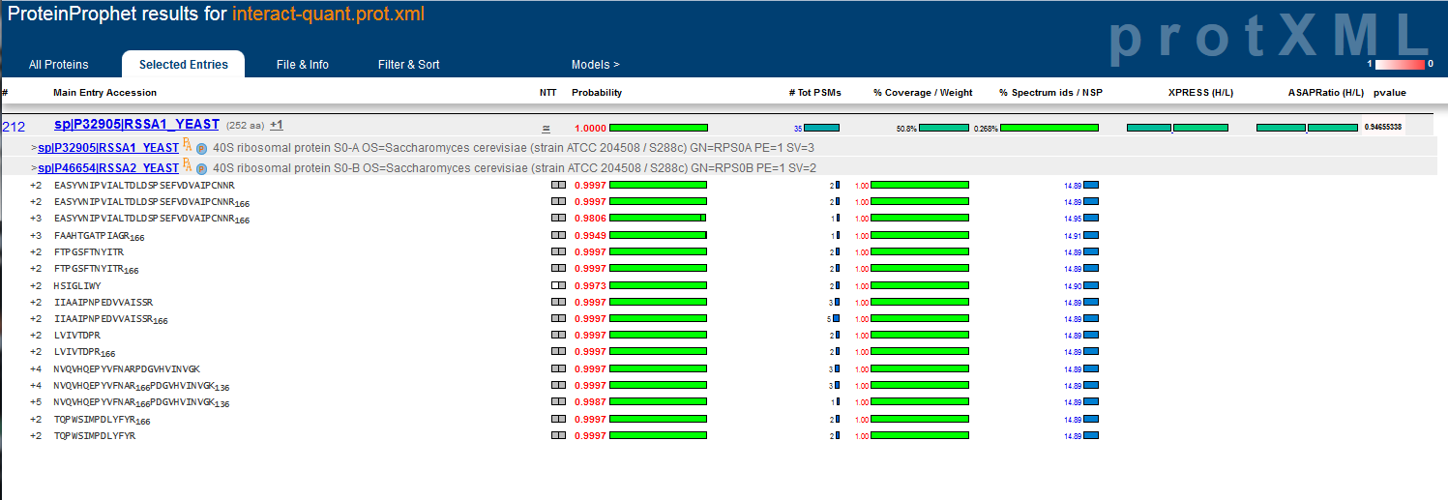
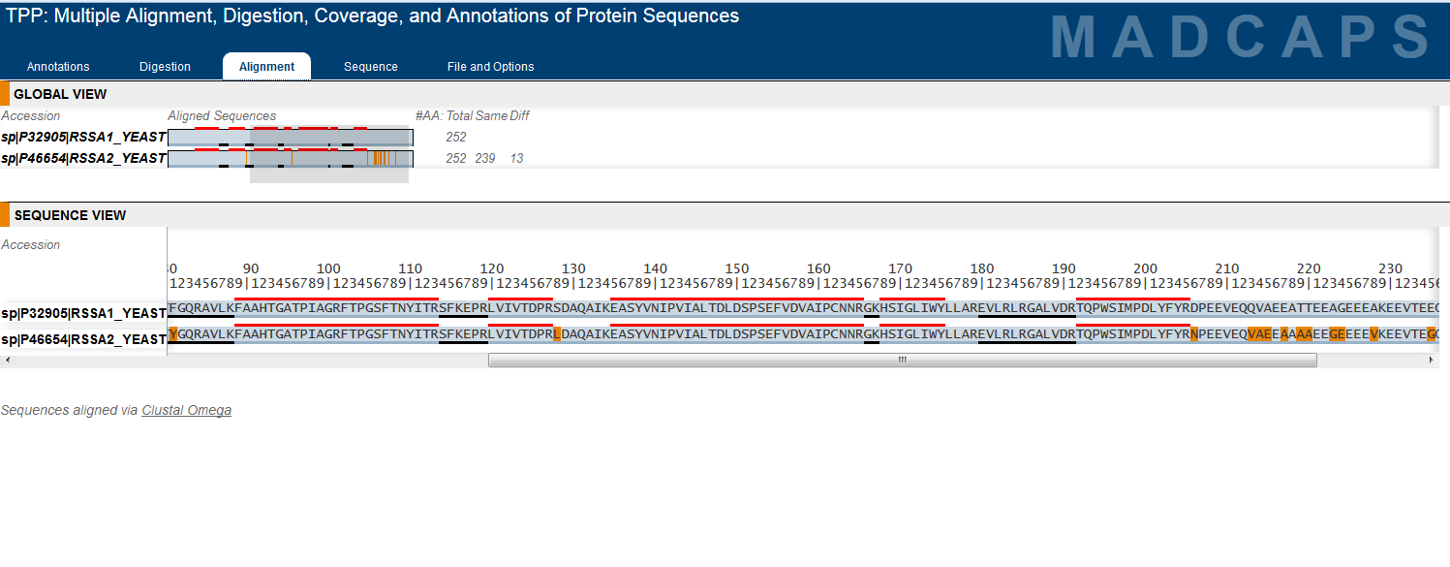
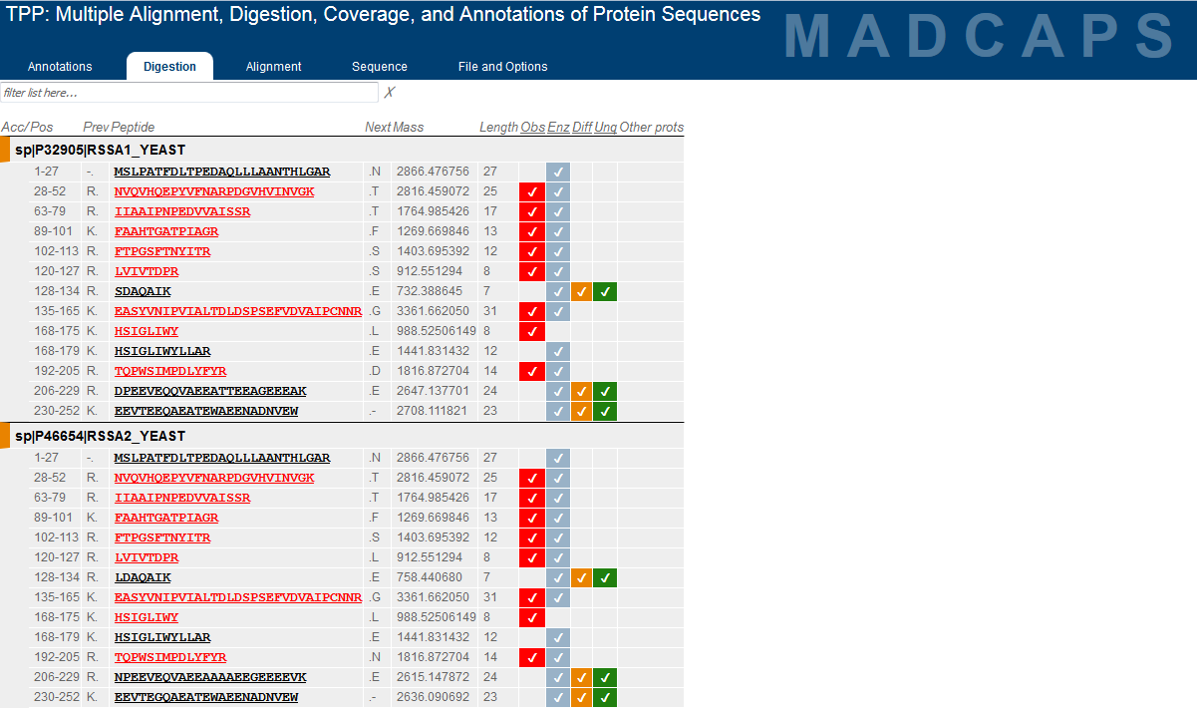
Development of the TPP was most recently supported by the National Institute Of General Medical Sciences of the National Institutes of Health under award number R01GM087221.
- ProteoMapper
- A set of tools to index and map peptides to proteome databases, including PEFF modifications